Tandem mass spectrometry
While enzymes, such as trypsin, can be used to cleave proteins and peptides at specific amino acid linkages, tandem mass spectrometry, we can also fragment peptides inside of a mass spectrometer to obtain additional information. In these experiments, protein mixtures are first digested with enzymes such as trypsinthen separated by one or more chromatography steps, and then electrosprayed into a tandem mass spectrometry spectrometer. Fragmentation requires that some energy be added to the system.
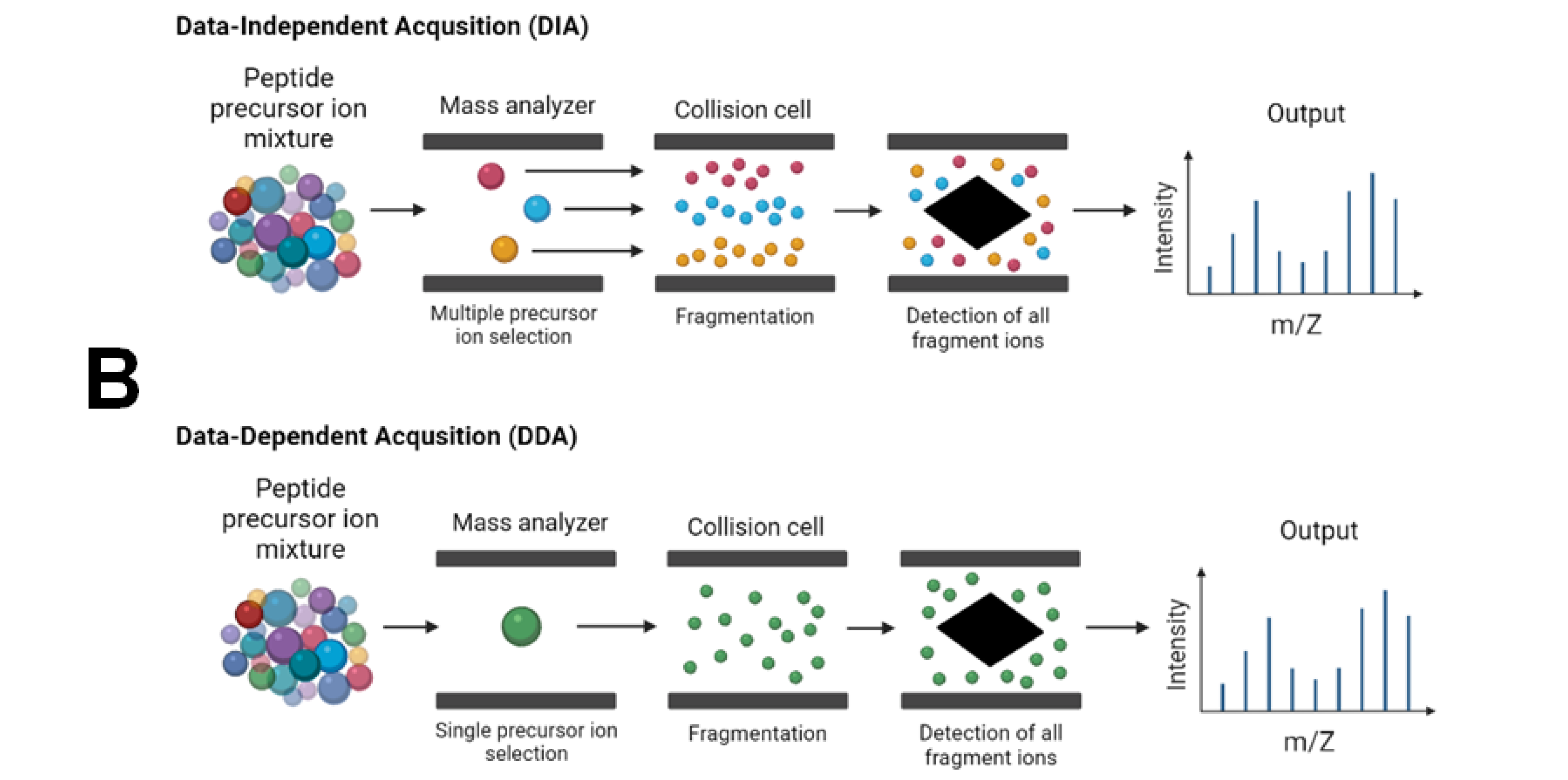
The fragments then reveal aspects of the chemical structure of the precursor ion. The following scheme explains how Tandem MS works. The selection-fragmentation-detection sequence can be further extended to the first-generation product ions. For example, selected product ions generated in MS2 can be further fragmented to produce another group of product ions MS3 and so on. Since Tandem MS involves three distinct steps of selection-fragmentation-detection, the separation of these three steps can be realized in space or in time. Three Quadrupoles Quad 1, Quad 2, and Quad 3 are lined up in a row. Precursor ions are selected in Quad 1 and sent to Quad 2 for dissociation fragmentation.
Tandem mass spectrometry
Federal government websites often end in. The site is secure. These methods allow identification of the mass of a protein or a peptide as intact molecules or the identification of a protein through peptide-mass fingerprinting generated upon enzymatic digestion. Furthermore, tandem mass spectrometry also allows the identification of post-translational modifications PTMs of proteins and peptides. Proteomics approaches have been employed in the last few decades for detecting and discriminating the early stages of diseases and for precise diagnoses to allow quick medical decisions and, consequently, to reduce mortality in various pathologies [ 3 ]. MS is a powerful analytical technique that is used to identify unknown compounds and to quantify known compounds [ 4 ]. Mass spectrometry provides information concerning the molecular structure, atomic mass of whole molecules, molecular fragments, and atoms [ 5 ]. Along with the well-known classical applications in biomarkers discovery, verification, and validation [ 8 ], MS-based diagnostic methods are involved today in the rapid and accurate identification of microbes [ 9 , 10 ], newborn screening [ 11 ], and quantification of therapeutic drugs [ 2 ]. MS is a powerful technique that detects the protein modifications in advanced personalized medicine, and the latest improvements in MS sensitivity and resolution allow the identification of new classes of tumor-specific proteoforms, including PTMs and variants originating from genomic and epigenomic aberrations [ 12 ]. A simple MS analysis is useful for the determination of proteins and peptide molecular weights by the detection of their molecular weight-related ions.
BioMed Res. During method development, acceptance criteria are established for the quality control and once the method is in production, the patient samples can only tandem mass spectrometry reported if the quality control samples pass these criteria.
In a tandem mass spectrometer, ions are formed in the ion source and separated by mass-to-charge ratio in the first stage of mass spectrometry MS1. Ions of a particular mass-to-charge ratio precursor ions are selected and fragment ions product ions are created by collision-induced dissociation, ion-molecule reaction, photodissociation, or other process. The resulting ions are then separated and detected in a second stage of mass spectrometry MS2. For tandem mass spectrometry in space, the different elements are often noted in shorthand. Multiple stages of mass analysis separation can be accomplished with individual mass spectrometer elements separated in space or using a single mass spectrometer with the MS steps separated in time. In tandem mass spectrometry in space , the separation elements are physically separated and distinct, although there is a physical connection between the elements to maintain high vacuum.
The fragments then reveal aspects of the chemical structure of the precursor ion. The following scheme explains how Tandem MS works. The selection-fragmentation-detection sequence can be further extended to the first-generation product ions. For example, selected product ions generated in MS2 can be further fragmented to produce another group of product ions MS3 and so on. Since Tandem MS involves three distinct steps of selection-fragmentation-detection, the separation of these three steps can be realized in space or in time.
Tandem mass spectrometry
Federal government websites often end in. The site is secure. Mass spectrometry is a powerful technique for chemical analysis that is used to identify unknown compounds, to quantify known compounds, and to elucidate molecular structure. It measures masses correspond to molecular structure and atomic composition of parent molecule and hence allows determination and elucidation of molecular structure [ 1 ]. Now the pertinent question comes to mind that why mass spectrometry? It may also be used for quantitation of molecular species. Mass spectrometry also provides valuable information to a wide range of professionals: chemists, biologists, physicians, astronomers, environmental health specialists. Tandem mass spectrometer is of many different types—each has different advantages, draw-backs and applications. All consist of four major sections linked together inlet—ionization source—analyser—detector. All sections are usually maintained under high vacuum and the functions of instrument control, sample acquisition and data processing are under computer control.
Eft warehouse 4
December Weerakoon H. Anal Chem. Qiu C. Zhang Y. Often, the ionization process is sufficiently violent to leave the resulting ions with sufficient internal energy to fragment within the mass spectrometer. Specifically, SID has been applied to the study of energetics and the kinetics of gas-phase fragmentation within an ICR instrument. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Thus, along with MSUD, PKU and homocystinuria, state programs can now test for argininosuccinic aciduria, citrullinemia and tyrosinemia [ 7 ]. It was invented by Donald F. Y: VCH Publishers.
Federal government websites often end in.
Article Google Scholar Shi, J. Federal government websites often end in. Article Google Scholar Polson, C. Fu, Q. Nature Reviews Methods Primers thanks Karl Storbeck, Joshua Hayden and the other, anonymous, reviewer s for their contribution to the peer review of this work. The liquid chromatography eluent carrying the analyte is introduced into the source of the mass spectrometer, where gas phase ions are produced. The numbers indicate the site of the sugar residue: y, z, b, and c ions are fragments due to glycosidic cleavages cutting glycosidic bonds holding two adjacent sugar residues , whereas a and x ions result from cross-ring cleavage. Casas-Vila N. Evaluation of sample preparation methods for mass spectrometry-based proteomic analysis of barley leaves. However, if the gradient time is extended to , , or min, then more proteins will be identified. The sample to be examined is essentially sorted and weighed in the first mass spectrometer, then broken into pieces in the collision cell, and a piece or pieces sorted and weighed in the second mass spectrometer. Christians, U. I would not like to post my testimonial.

0 thoughts on “Tandem mass spectrometry”