Scispacy
This repository contains custom pipes and models related to using spaCy for scientific documents. In particular, scispacy, there scispacy a custom tokenizer that adds tokenization rules on top of spaCy's rule-based tokenizer, a POS tagger and syntactic parser trained on biomedical data and an entity span detection model, scispacy. Separately, there are also NER models for more specific tasks.
A beginner's guide to using Named-Entity Recognition for data extraction from biomedical literature. This code walks you through the installation and usage of scispaCy for natural language processing. For our example, we use data from CORD, a large collection of articles about the Covid pandemic. It is a very powerful tool, especially for named entity recognition NER , but it can be somewhat confusing to understand. The goal of this code is to show scispaCy in easy to understand terms. I hope it makes navigating the world of entity extraction a little easier.
Scispacy
Released: Feb 20, View statistics for this project via Libraries. Author: Allen Institute for Artificial Intelligence. Tags bioinformatics, nlp, spacy, SpaCy, biomedical. Mar 8, Sep 30, Apr 29, Sep 7, Mar 10, Feb 12, Oct 16, Jul 8, Oct 22, Aug 22, Jun 3,
Sep 30, Installation Installing scispacy requires two steps: installing the library and intalling the models, scispacy.
.
In its most basic form a spaCy application can be very short, but a lot of processing steps take place, and a lot more information is contained within the doc object. If your result is a shorter list of pipeline components then you are likely not using the most recent version of spaCy. Here is some of the information that is available from the nlp object:. There are three main types of text models used in NLP: rules-based models, statistics-based models, and neural network-based models. The second two of these both fall into the category of machine learning, but nerual networks or deep learning required a lot more RAM and processing power. On the other hand, they can be very effective, and are increasingly the norm for natural language processing. A variety of different statistical and neural network models can be imported into the spaCy pipeline. They generally. Decisions are about parts of speech, named entities, etc. There are a variety of different types of models available in spaCy—and related projects —and are installed in spaCy as Python packages.
Scispacy
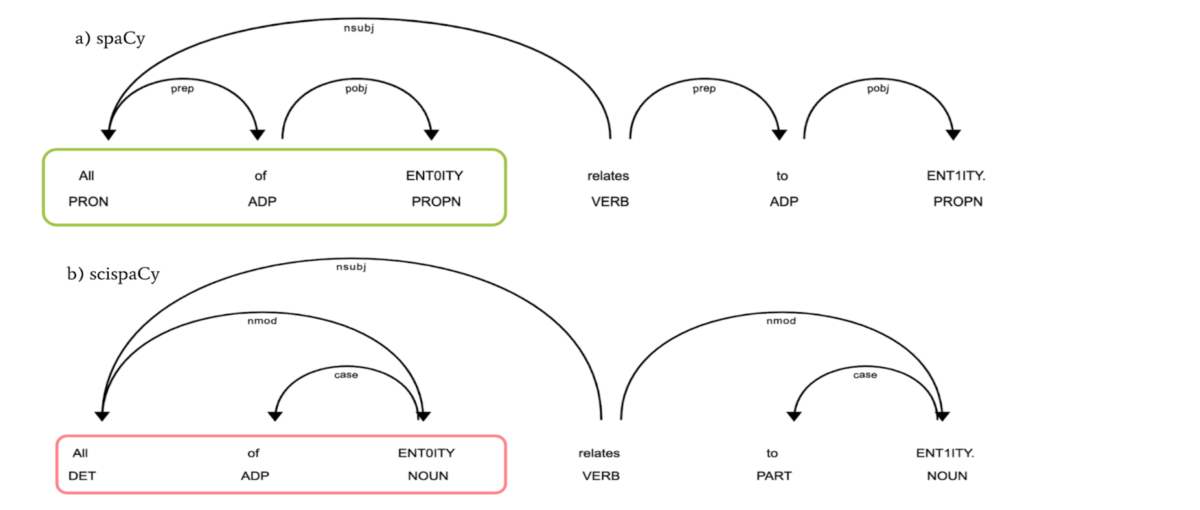
Despite recent advances in natural language processing, many statistical models for processing text perform extremely poorly under domain shift. Processing biomedical and clinical text is a critically important application area of natural language processing, for which there are few robust, practical, publicly available models. We detail the performance of two packages of models released in scispaCy and demonstrate their robustness on several tasks and datasets. The publication rate in the medical and biomedical sciences is growing at an exponential rate Bornmann and Mutz, The information overload problem is widespread across academia, but is particularly apparent in the biomedical sciences, where individual papers may contain specific discoveries relating to a dizzying variety of genes, drugs, and proteins. In order to cope with the sheer volume of new scientific knowledge, there have been many attempts to automate the process of extracting entities, relations, protein interactions and other structured knowledge from scientific papers Wei et al. Although there exists a wealth of tools for processing biomedical text, many focus primarily on named entity recognition and disambiguation. In this paper, we introduce scispaCy, a specialized NLP library for processing biomedical texts which builds on the robust spaCy library, 1 1 1 spacy. Specifically, we:. Benchmark 9 named entity recognition models for more specific entity extraction applications demonstrating competitive performance when compared to strong baselines.
Structube return policy
View all files. Latest commit History 8 Commits. Tags bioinformatics, nlp, spacy, SpaCy, biomedical. Jul 8, We detail the performance of two packages of models released in scispaCy and demonstrate their robustness on several tasks and datasets. Please try enabling it if you encounter problems. Close Hashes for scispacy You switched accounts on another tab or window. Then, we display our results. This section expands the previous code to loop over the entire csv file and continually grab the text we want and use NER to grab the entities and their attributes. Be patient! Oct 22, You will need to activate the Conda environment in each terminal in which you want to use scispaCy. A beginner's guide to using Named-Entity Recognition for data extraction from biomedical literature.
The goal of clinspacy is to perform biomedical named entity recognition, Unified Medical Language System UMLS concept mapping, and negation detection using the Python spaCy, scispacy, and medspacy packages. Restarting your R session should resolve the issue. Initiating clinspacy is optional.
Report repository. This class sets the. Search PyPI Search. Report repository. Download the file for your platform. If you're not sure which to choose, learn more about installing packages. Additionally, please indicate which version and model of ScispaCy you used so that your research can be reproduced. Example Helper Method. Latest commit. Newer version available 0. The models are installed using their URLs, found here. A beginner's guide to using Named-Entity Recognition for data extraction from biomedical literature.

0 thoughts on “Scispacy”