Riboswitch
Federal government websites often end in.
Riboswitches are specific components of an mRNA molecule that regulates gene expression. The riboswitch is a part of an mRNA molecule that can bind and target small target molecules. An mRNA molecule may contain a riboswitch that directly regulates its own expression. The riboswitch displays the ability to regulate RNA by responding to concentrations of its target molecule. Hence, the existence of RNA molecules provide evidence to the RNA world hypothesis that RNA molecules were the original molecules, and that proteins developed later in evolution. Riboswitches are found in bacteria, plants, and certain types of fungi. The various mechanisms by which riboswitches function can be divided into two major parts including an aptamer and an expression platform.
Riboswitch
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Cell-free genetically encoded biosensors have been developed to detect small molecules and nucleic acids, but they have yet to be reliably engineered to detect proteins. Here we develop an automated platform to convert protein-binding RNA aptamers into riboswitch sensors that operate within low-cost cell-free assays. The riboswitch sensors regulate output expression levels by up to fold with input protein concentrations within the human serum range. We identify two distinct mechanisms governing riboswitch-mediated regulation of translation rates and leverage computational analysis to refine the protein-binding aptamer regions, improving design accuracy. Overall, we expand the cell-free sensor toolbox and demonstrate how computational design is used to develop protein-sensing riboswitches with future applications as low-cost medical diagnostics. Samuel I. Gould, Alexandra N. Wuest, … Francisco J.
This folding process can be monitored by a range of methods including classical approaches riboswitch use temperature and chemical denaturants to measure relative stability of structural elements, riboswitch, and more recently developed single molecule techniques that monitor folding transitions in real time. Since riboswitches are an effective method of controlling gene expression in natural organisms, there has been interest in engineering artificial riboswitches [42] [43] [44] for industrial and riboswitch applications such as gene therapy. Sproston, riboswitch, N.
This page has been archived and is no longer updated. Every living organism must be able to sense environmental stimuli and convert these input signals into appropriate cellular responses. Most of these responses are mediated by transcription factors that bind DNA and coordinate the activity of RNA polymerase or of proteins that elicit allosteric effects on their regulatory targets. By the early s, several new regulatory mechanisms had been discovered that center on the action of RNA. Arnaud et al.
Federal government websites often end in. The site is secure. Preview improvements coming to the PMC website in October Learn More or Try it out now. A growing collection of bacterial riboswitch classes is being discovered that sense central metabolites, coenzymes, and signaling molecules.
Riboswitch
This page has been archived and is no longer updated. Every living organism must be able to sense environmental stimuli and convert these input signals into appropriate cellular responses. Most of these responses are mediated by transcription factors that bind DNA and coordinate the activity of RNA polymerase or of proteins that elicit allosteric effects on their regulatory targets. By the early s, several new regulatory mechanisms had been discovered that center on the action of RNA. Arnaud et al. Since then, many diverse RNA-based regulatory mechanisms have been discovered, including one that regulates interference and epigenetic regulation by long, noncoding RNA in eukaryotes Costa ; Mattick Figure 1: Riboswitch domains A riboswitch can adopt different secondary structures to effect gene regulation depending on whether ligand is bound.
Hubporner
Shaw, W. Comparative genomics reveals candidate structured RNAs from bacteria, archaea, and their metagenomes. Trends Biochem Sci 40 : — Structural schematics were partly made using Forna diagrams The S MK box riboswitch is found only in the order Lactobacillales. However, monitoring similar kinetic processes that involve ribosome docking to a riboswitch-controlled RBS, or spliceosomal particles choosing splice sites whose availabilities are controlled by riboswitch folding, seem daunting. Genome Biol 11 : R Genome Biology 8 , Interaction between the loop and the receptor serves to anchor the two helices together in a side-by-side arrangement, facilitating parallel packing. Wuest, … Francisco J. Therefore, controlling ribosome access must involve alternative folding of structures apart from the aptamer that either hide or openly display the RBS. Addition of the appropriate purine nucleobase induces structure in the three-way junction, as evidenced both by the appearance of new peaks in NMR spectra Noeske et al. Trends in Biochemical Sciences 29 , 11—17
Federal government websites often end in. The site is secure.
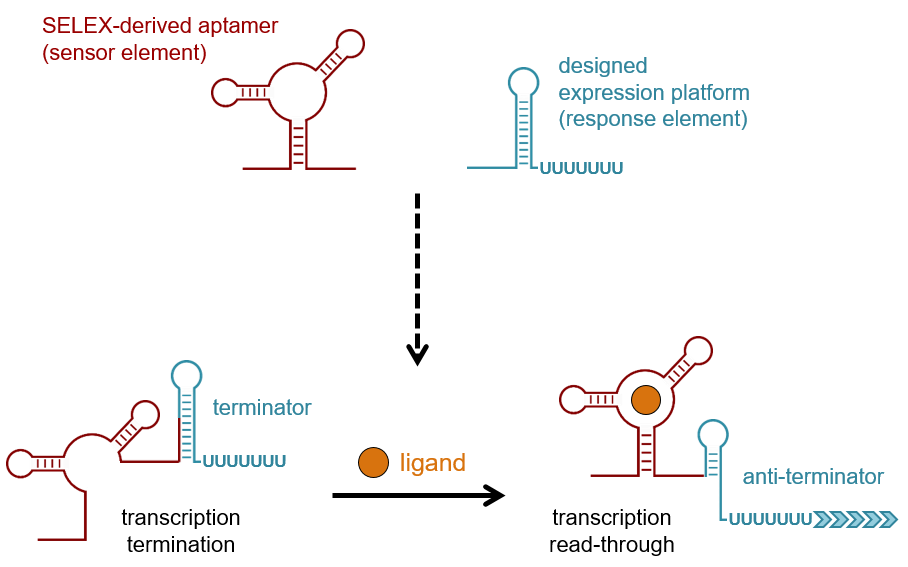
YkoK leader : Secondary structure for the riboswitch marked up by sequence conservation. Temperature dependence of RNA synthesis parameters in Escherichia coli. This is because the undiscovered classes tend to be rarer fewer number of representatives than those discovered previously McCown et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. In most instances, binding of a target ligand to the aptamer domain of the riboswitch triggers changes in the folding pattern of the expression platform Fig. Riboswitches as versatile gene control elements. Annu Rev Microbiol 70 : — A second type has the RBS located at some distance downstream of the aptamer structure.

I think, that you are not right. Let's discuss.
I join. And I have faced it.
Remove everything, that a theme does not concern.