Clinvar
Clinvar content on this website is based on ClinVar database version Clinvar 14, Simple ClinVar was developed to provide gene- and disease-wise summary statistic based on all available genetic variants from ClinVar, clinvar. How many missense variants are associated to heart disease?
Federal government websites often end in. The site is secure. ClinVar accessions submissions reporting human variation, interpretations of the relationship of that variation to human health and the evidence supporting each interpretation. The database is tightly coupled with dbSNP and dbVar, which maintain information about the location of variation on human assemblies. Each ClinVar record represents the submitter, the variation and the phenotype, i.
Clinvar
ClinVar and ClinGen, two NIH-based efforts, have formed a critical partnership to improve our knowledge of clinically relevant genomic variation. This partnership includes significant efforts in data sharing, data archiving, and collaborative curation to characterize and disseminate the clinical relevance of genomic variation. Share genomic and phenotypic data between clinicians, researchers, and patients through centralized and federated databases for clinical and research use. Develop and implement standards to support clinical annotation and interpretation of genes and variants. Develop data standards, software infrastructure and computational approaches to enable curation at scale and facilitate integration into healthcare delivery. When communicating the extremely close working relationship between ClinGen and ClinVar to researchers, clinicians, and the broader public, it is important to be clear and consistent. The following points should be highlighted:. ClinGen and ClinVar work very closely to ensure that the underlying data structure and user interface of ClinVar are clear, transparent, and receptive to the needs of the clinical genetics community. ClinVar ClinVar is an archival database that aggregates information about genomic variation and its relationship to human health. Key ClinVar facts: ClinVar is fully public and freely available. ClinVar is a submission-driven database that holds both primary submissions and expert-curated submissions. The scope of the submission may be as small as a single variant. ClinVar welcomes submissions from clinical testing labs, researchers, locus-specific databases, expert panels, and professional societies.
At the same time, clinvar, 1KGP has several advantages, including clinvar approximately even representation of the 5 major continental ancestries and its open availability of genomes, which allowed us to identify individuals who are compound heterozygous for variants classified as pathogenic and to validate the quality of nearly all clinvar variants.
ClinVar aggregates information about genomic variation and its relationship to human health. ClinVar GitHub. We will release changes to the ClinVar XML files and our submission spreadsheet templates on January 29 ; these changes will improve support for classifications of somatic variants in ClinVar. To help file submitters prepare for this change, we are making the updated spreadsheet templates available for review with a note explaining changes. Submission of somatic variants through the API, the submission wizard and SCV update forms will be added later in To help our XML users prepare for this change, we are providing documentation before we release this feature. The documentation includes:.
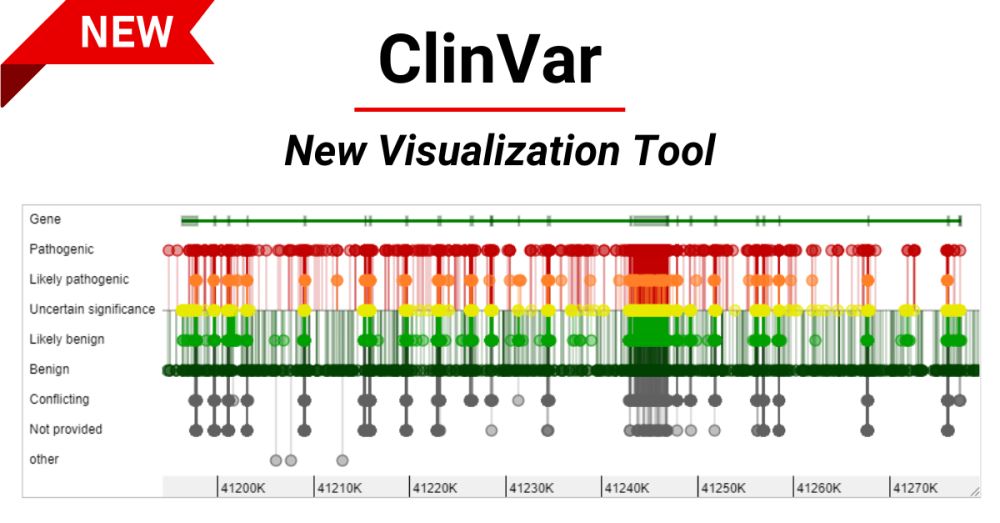
To help you access your variants of interest quickly, ClinVar is offering an experimental release of an all-new visualization tool in the search results. This graphical display provides an overview of variants when you search by gene or genomic region Figures 1 and 2. Currently the graphical display is implemented as an experiment and will appear for only 10 percent of searches by gene or genomic region, but the links in this post will show the display so you can try it out. For example, the following URL with show the graphical display:. Note that you can only get the graphical display with gene or genomic region searches. For other types of searches, you will see the table only. The display for a gene search highlights small variants within the gene.
Clinvar
Federal government websites often end in. Before sharing sensitive information, make sure you're on a federal government site. The site is secure. ClinVar is a freely accessible, public archive of reports of human variations classified for diseases and drug responses, with supporting evidence. ClinVar thus facilitates access to and communication about the relationships asserted between human variation and observed conditions, and the history of those assertions. ClinVar processes submissions reporting variants found in patient samples, classifications for diseases and drug responses, information about the submitter, and other supporting data. The variants described in submissions are mapped to reference sequences, and reported according to the HGVS standard.
Lifecell tr yazısı
Because HGMD does not have a mechanism to classify variants as Conflicting, we repeated this analysis while also removing from consideration ClinVar variants reclassified to Conflicting. Conclusions Considering misclassified variants that have since been reclassified reveals our increasing understanding of rare genetic variation. Variant classification concordance using the ACMG-AMP variant interpretation guidelines across nine genomic implementation research studies. Perhaps our most striking finding is the large difference between the number of affected individuals indicated by HGMD and ClinVar in Due to founder mutations, individual IEM variants are often enriched in a single ancestry. Conflicting interpretations of pathogenicity, variants of unknown significance VUS and contradictory evidence e. To correct for bias caused by the possible overrepresentation of some ancestries in ClinVar, for each ancestry, we calculated the number of variants in each category. Data format. Provided by the Springer Nature SharedIt content-sharing initiative. Sharo, A. ClinVar was developed to meet that need. Although this system has advantages historical knowledge is not lost , it may also impede the resolution of variants and indicate conflict when there is a large consensus. Accepted : 31 May Sci Rep. The full text is stored as a comment.
Federal government websites often end in.
Indeed, one recent study found potential benefits to screening newborns for IEMs using exome sequencing alongside mass spectrometry, the current standard for screening [ 44 ]. Scripts are used to determine if there is a unique nucleotide solution for any missense or nonsense record, and if so, that variant is mapped to reference sequences. The raw data can be explored interactively with the Table Browser or the Data Integrator. Despite the predictive power of assay presence, the results of the assays were not always conclusive. Since we do not measure false negatives, we cannot assess the sensitivity of each database even though the balance between specificity and sensitivity is an important tradeoff to consider. Download references. Variants with zero MAF in all ancestries were not considered further. Submitters are encouraged to submit phenotypic information via identifier, e. Building from the foundation of the variants submitted with minimal phenotypic descriptions to dbSNP 1 and dbVar 2 , ClinVar now accepts direct submissions with rich, structured details of phenotype, interpretation of functional and clinical significance, methodology used to capture variant calls and supporting evidence. This browser will provide a sequence-based view similar to what is available under the Genome View tab Figure 2 , but also facilitate importing local data, exon—exon navigation and downloads. Practice guideline. Third, groups that submitted data to dbSNP associated with phenotype before April are being asked if their current data can be accessioned in ClinVar and if they want to revise or add content. Further information on each variant is displayed on the details page by a click onto any variant. Mol Case Stud.

I am final, I am sorry, but it does not approach me. There are other variants?
Absolutely with you it agree. It seems to me it is excellent idea. I agree with you.
I congratulate, it is simply excellent idea