Clin var
Federal government websites often end in.
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Automated variant filtering is an essential part of diagnostic genome-wide sequencing but may generate false negative results.
Clin var
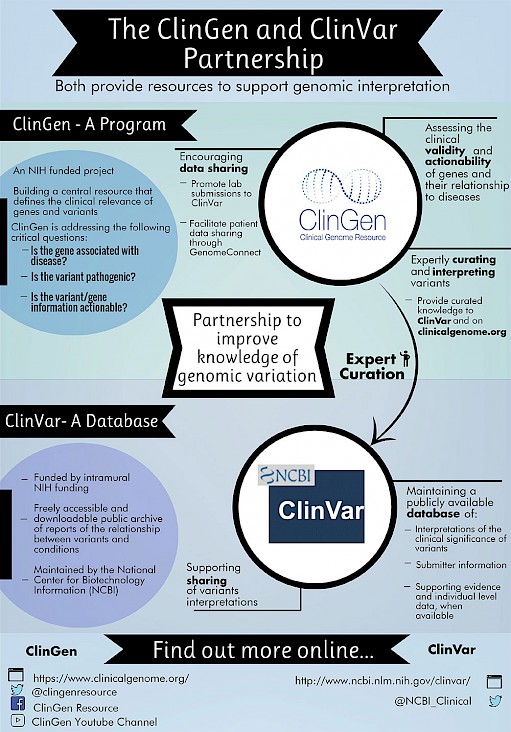
Interpretations of the clinical significance of variants are submitted by clinical testing laboratories, research laboratories, expert panels and other groups. ClinVar aggregates data by variant-disease pairs, and by variant or set of variants. Data aggregated by variant are accessible on the website, in an improved set of variant call format files and as a new comprehensive XML report. ClinVar recently started accepting submissions that are focused primarily on providing phenotypic information for individuals who have had genetic testing. ClinVar continues to make improvements to its search and retrieval functions. Several new fields are now indexed for more precise searching, and filters allow the user to narrow down a large set of search results. ClinVar 1 , 2 is a freely available, public archive of human genetic variants and interpretations of their significance to disease. Assertions of the clinical significance of a variant or set of variants are submitted to ClinVar by clinical testing laboratories, research laboratories, locus-specific databases, expert panels and other groups. Submissions include a description of the variant s ; the condition for which the variant was interpreted; the interpretation of the clinical significance of the variant, with the option to provide mode of inheritance; and evidence for that interpretation. ClinVar aggregates submissions based both on the variant and the variant-condition pair, and calculates an aggregate interpretation to indicate whether there is consensus or disagreement among submitters for an interpretation.
At present, clin var, most structured data are reports of number of individuals, in which non-somatic variation was observed, sometimes with indication of number of families.
Genome Medicine volume 15 , Article number: 51 Cite this article. Metrics details. Curated databases of genetic variants assist clinicians and researchers in interpreting genetic variation. Yet, these databases contain some misclassified variants. It is unclear whether variant misclassification is abating as these databases rapidly grow and implement new guidelines. Using archives of ClinVar and HGMD, we investigated how variant misclassification has changed over 6 years, across different ancestry groups. We considered inborn errors of metabolism IEMs screened in newborns as a model system because these disorders are often highly penetrant with neonatal phenotypes.
Federal government websites often end in. Before sharing sensitive information, make sure you're on a federal government site. The site is secure. Go to the search box in the gray area at the top of the page. Just type your search term and click on the Search button to the right of the search box. ClinVar can be searched with terms like:. In other words, when you enter a term or phrase of interest in the query box, that term or phrase will be processed to retrieve records that contain or have some relationship to the word s you entered.
Clin var
Federal government websites often end in. Before sharing sensitive information, make sure you're on a federal government site. The site is secure. ClinVar is a freely accessible, public archive of reports of human variations classified for diseases and drug responses, with supporting evidence. ClinVar thus facilitates access to and communication about the relationships asserted between human variation and observed conditions, and the history of those assertions. ClinVar processes submissions reporting variants found in patient samples, classifications for diseases and drug responses, information about the submitter, and other supporting data. The variants described in submissions are mapped to reference sequences, and reported according to the HGVS standard. ClinVar presents the data on the website for interactive users, and on the FTP site and by API for those wishing to use ClinVar programmatically in daily workflows and other local applications. ClinVar works in collaboration with interested organizations to meet the needs of the medical genetics community as efficiently and effectively as possible. ClinVar supports submissions of differing levels of complexity.
Despicable me stuart
Out of variants classified as DM or DM? International cooperation to enable the diagnosis of all rare genetic diseases. Although we cannot be certain that no individual in 1KGP has a screened IEM, this metric is a proxy for the false-positive rate of each database. Develop and implement standards to support clinical annotation and interpretation of genes and variants. Evolution of the Sequence Ontology terms and relationships. This resulted in a complex aggregation of data which was complicated to parse. A survey assessing adoption of the ACMG-AMP guidelines for interpreting sequence variants and identification of areas for continued improvement. View author publications. About this article. Lee , George R. As of , Full ClinVar variants indicate 0. Informed consent was obtained from all subjects.
.
ClinVar continues to make improvements to its search and retrieval functions. By , eight of these variants were classified as Conflicting. Crystal npj Genomic Medicine Copyright Published by Oxford University Press Revised : 14 October More information on ClinVar is available at www. Until recently, variants were primarily classified in locus-specific databases LSDBs that typically collected variants in a single gene. Instead, we examined the evidence for pathogenicity of the 19 Select variants identified in an inferred pathogenic genotype in , , or Anyone you share the following link with will be able to read this content:. Zenith Maddipatla. As of , there was a single 1KGP indicated affected individual. A record retrieved by an outdated ID provides a link to the current record. At present September , this display corresponds to content of an RCV accession. ClinGen relies on ClinVar as a source for existing data on variants, which are submitted to ClinVar from diverse sources. If submitters disagree on the interpretation of the clinical significance of any variation, the aggregate record is marked as having conflicts.

It is remarkable, it is very valuable piece