Allosteric
In biochemistryallosteric regulation or allosteric control is the regulation of an enzyme by binding an effector molecule at a site other than the enzyme's active site, allosteric. The site to which the effector binds is termed the allosteric site or regulatory site. Allosteric regulations are a natural example of allosteric loops, such as feedback from downstream products or feedforward from allosteric substrates, allosteric. Long-range allostery is especially important in cell signaling.
Federal government websites often end in. The site is secure. Allosteric drugs are currently receiving increased attention in drug discovery because drugs that target allosteric sites can provide important advantages over the corresponding orthosteric drugs including specific subtype selectivity within receptor families. Consequently, targeting allosteric sites, instead of orthosteric sites, can reduce drug-related side effects and toxicity. On the down side, allosteric drug discovery can be more challenging than traditional orthosteric drug discovery due to difficulties associated with determining the locations of allosteric sites and designing drugs based on these sites and the need for the allosteric effects to propagate through the structure, reach the ligand binding site and elicit a conformational change. These tools may be particularly useful for allosteric drug discovery. Allostery, which is also known as allosteric regulation, is an essential biological phenomenon that plays significant roles in signal transduction pathways, metabolic processes, and genomic transcription [ 1 , 2 ].
Allosteric
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Allosteric communication between distant sites in proteins is central to biological regulation but still poorly characterized, limiting understanding, engineering and drug development 1 , 2 , 3 , 4 , 5 , 6. An important reason for this is the lack of methods to comprehensively quantify allostery in diverse proteins. Here we address this shortcoming and present a method that uses deep mutational scanning to globally map allostery. The approach uses an efficient experimental design to infer en masse the causal biophysical effects of mutations by quantifying multiple molecular phenotypes—here we examine binding and protein abundance—in multiple genetic backgrounds and fitting thermodynamic models using neural networks. We apply the approach to two of the most common protein interaction domains found in humans, an SH3 domain and a PDZ domain, to produce comprehensive atlases of allosteric communication. Mutations are more likely to be allosteric closer to binding interfaces, at glycine residues and at specific residues connecting to an opposite surface within the PDZ domain. This general approach of quantifying mutational effects for multiple molecular phenotypes and in multiple genetic backgrounds should enable the energetic and allosteric landscapes of many proteins to be rapidly and comprehensively mapped. This is a preview of subscription content, access via your institution. Guarnera, E.
Popular in Wordplay See All. ADS Google Scholar.
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Allostery in proteins influences various biological processes such as regulation of gene transcription and activities of enzymes and cell signaling. Computational approaches for analysis of allosteric coupling provide inexpensive opportunities to predict mutations and to design small-molecule agents to control protein function and cellular activity. We develop a computationally efficient network-based method, Ohm, to identify and characterize allosteric communication networks within proteins.
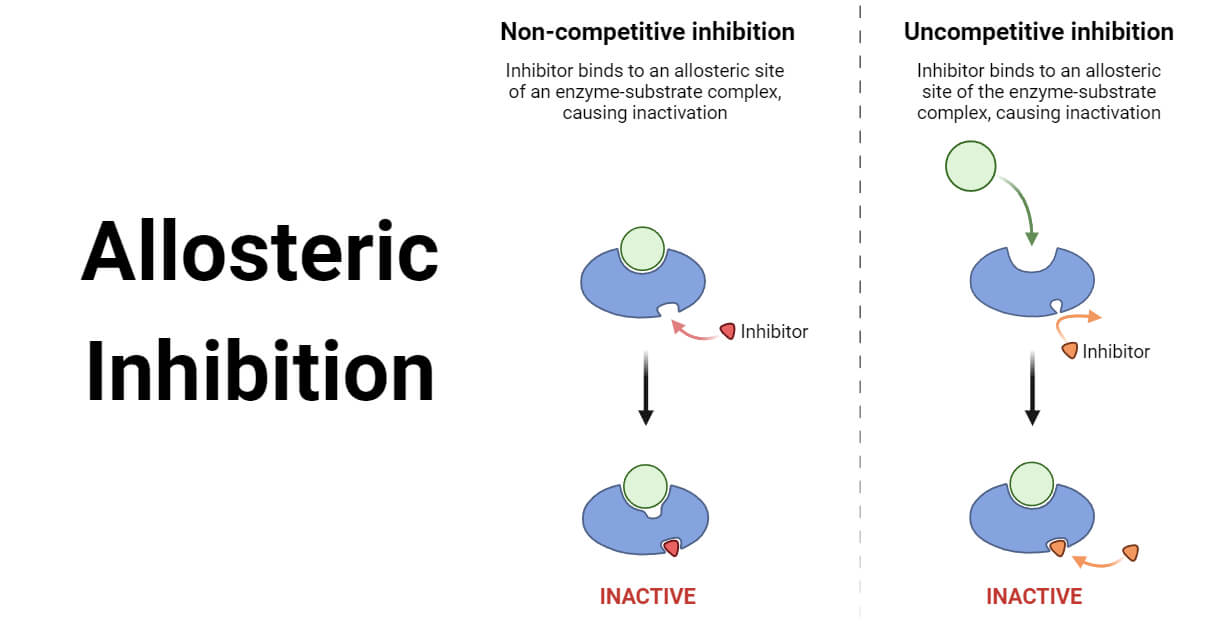
In biochemistry , allosteric regulation or allosteric control is the regulation of an enzyme by binding an effector molecule at a site other than the enzyme's active site. The site to which the effector binds is termed the allosteric site or regulatory site. Allosteric regulations are a natural example of control loops, such as feedback from downstream products or feedforward from upstream substrates. Long-range allostery is especially important in cell signaling. This is in reference to the fact that the regulatory site of an allosteric protein is physically distinct from its active site. The two models differ most in their assumptions about subunit interaction and the preexistence of both states. For proteins in which subunits exist in more than two conformations , the allostery landscape model described by Cuendet, Weinstein, and LeVine, [6] can be used. Allosteric regulation may be facilitated by the evolution of large-scale, low-energy conformational changes, which enables long-range allosteric interaction between distant binding sites.
Allosteric
In pharmacology and biochemistry , allosteric modulators are a group of substances that bind to a receptor to change that receptor's response to stimuli. Some of them, like benzodiazepines or alcoholic beverages , function as psychoactive drugs. Modulators and agonists can both be called receptor ligands.
A night with loona 2
Introduction Allostery, which is also known as allosteric regulation, is an essential biological phenomenon that plays significant roles in signal transduction pathways, metabolic processes, and genomic transcription [ 1 , 2 ]. Article Talk. PMID Allosteric proteins and cellular control systems. The propagation is performed 10 4 times and the histogram of all paths is statistically evaluated and stored in S. Thus, all enzyme subunits do not necessitate the same conformation. Calosci, N. By definition, allostery involves the propagation of signals between sites in a protein structure through a network of residues 22 , 64 , Datta, D. Simonovic, M. Thus, users can predict cryptic allosteric sites with Allosite if representative conformations are chosen as inputs. View author publications. Supplementary Data 4. Nucleic Acids Research.
A significant portion of enzymes function such that their properties can be studied using the Michaelis-Menten equation. However, a particular class of enzymes exhibit kinetic properties that cannot be studied using the Michaelis-Menten equation.
Take the quiz. For proteins, the selection of an appropriate distance cutoff of contacts, the exclusion of backbone—backbone atom contacts between two sequence-adjacent residues, and the formula that converts contacts to probability all play crucial parts in computing the perturbation propagation probability matrix. We thank M. Non-regulatory allostery could comprise any other ions besides sodium calcium, magnesium, zinc , as well as other chemicals and possibly vitamins. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Each cluster is predicted as an allosteric hotspot. Thus, we recommend to use all residues on the active site for the unbound structure. Regions in dashed circles only appear in allosteric correlations predicted by Ohm. Annual Review of Biochemistry. Next, each of the residues in the active site is assigned 1. Mosca, R. Allosteric sites allow effectors to bind to the protein, often resulting in a conformational change involving protein dynamics.

In my opinion it is obvious. I recommend to look for the answer to your question in google.com